Data
Sexually Transmitted Infections Pipeline
Developed as part of the G-FINDER project, this pipeline captures critical products for three sexually transmitted infections (STIs) with a risk of antimicrobial resistance (AMR). This database covers drugs, vaccines, biologics, microbicides and diagnostics marketed and/or in active development from 2021 to 2024.
Table 1: Scope of pipeline
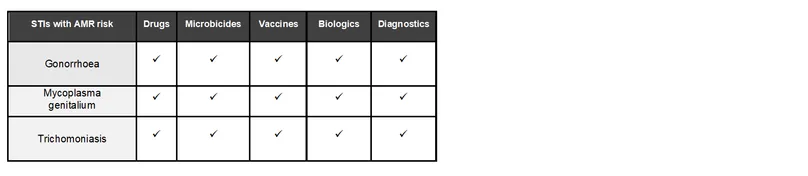
The pipeline we developed is candidate-focused (as opposed to clinical trial-focused) presenting a profile for each candidate identified. It focuses on active candidates, with evidence of activity between 2021 and 2024, and products (marketed, approved or adopted in clinical practice as gold standard – e.g. diagnostic tests).
This first iteration of the SRH pipeline focuses on sexually transmitted infections (STIs) with a risk of antimicrobial resistance (AMR). Based on the WHO’s evaluation, three STI pathogens are at risk of AMR: Neisseria gonorrhoeae, Mycoplasma genitalium and Trichomonas vaginalis.
The product categories included in the pipeline for these three STIs are drugs, biologics, vaccines, microbicides and diagnostics.
For inclusion in the pipeline dataset, medicine entries must:
- be drugs (small molecules intended for use in the cure, mitigation, treatment, or prevention of disease), biologics (animal or non-animal derived, immunoglobulins or non-immunoglobulin products) with no restrictions: they can be entirely new entities; existing/repurposed/label extensions; new formulations or dosing of existing/registered medicines.
- be preventive or therapeutic in nature – used to prevent the transmission of STIs, the acquisition/development of the infection or to eradicate the infection;
- have one or multiple indications related to either gonorrhoea, mycoplasma genitalium or trichomoniasis;
- either be in active discovery, pre-clinical or clinical development since 2021, or be approved and registered for clinical use and/or be used currently in clinical treatment (off-label), and;
- be applicable for use in LMIC context.
For inclusion in the pipeline dataset, diagnostic entries must:
- be predictive/screening and/or diagnostic tests, with no restrictions: biochemical serum markers; non-biomarker tests; or fixed multiparametric tests;
- have an indication or multiple indications related to gonorrhoea, mycoplasma genitalium or trichomoniasis; either for the diagnosis of the STI or for diagnosing antimicrobial resistance
- either be in active discovery, pre-clinical or clinical development since 2021, or be approved and registered for clinical use and/or be used currently in clinical treatment (off-label), and;
- be applicable for use in LMIC context.
For inclusion in the pipeline dataset, vaccines entries must:
- be preventive or therapeutic in nature – used to prevent the transmission of STIs, the acquisition/development of the infection or to clear the infection;
- have one or multiple indications related to either gonorrhoea, mycoplasma genitalium or trichomoniasis
- either be in active discovery, pre-clinical or clinical development since 2021, or be approved and registered for clinical use and/or be used currently in clinical treatment (off-label), and;
- be applicable for use in LMIC context.
For inclusion in the pipeline dataset, microbicide entries must:
- be topical microbicides intended to prevent transmission of N. gonorrhoea, M. genitalium or trichomonas vaginalis
- be suitable for use in LMICs (e.g. heat stable, easy to use)
- If microbicides are designed to prevent two or more STIs, or combined with contraceptives, they are considered multi-purpose preventive technologies (MPTs)
Exclusions:
- Dietary supplements are excluded;
- Predictive/diagnostic strategies, algorithms with no identifiable products; or that include risk models that comprise of multiple individual parameters/tests as a single candidate entry; (for example several individual biomarkers which diagnostic utility is evaluated separately and not as a combined test usable in the clinic);
- Clinical guidelines or criteria that are not a biomedical product are excluded.
The pipeline includes the following conditions and products:
- Gonorrhoea, Mycoplasma Genitalium and Trichomoniasis: drugs, biologics, vaccines, microbicides, diagnostics
All entries contain the same information. Data fields for each new candidate include all the following fields, where available and verifiable:
Table 2: Data fields

Data sources and approach
To build out comprehensive, candidate-driven pipeline datasets, we undertook a series of partially sequential, mutually reinforcing steps to build a picture of the candidates themselves, as well as the product development landscape for that SRH condition. Broadly (with more detail below), these steps are: (1) identify all candidates that are marketed or in development for each SRH health area and associated product type (e.g. Endometriosis diagnostics) and establish unique profiles for each with standardised classifications across a range of standard characteristics (2) gather information on that candidate’s clinical development and pre-clinical/clinical trial results, (3) research additional context around the product (e.g. development history, stakeholders, etc.), and (4) sense check our findings.
Step 1: Initial candidate and product identification.
We utilised various sources to uncover, identify and cross-check candidates or marketed products. Key sources include:
- Adis Insight– a leading medicines development database, which will give a comprehensive output of relevant pipeline drugs, biologics (and possibly devices) related to relevant SRH conditions. Adis Insight also returns detailed information on candidate deals, trials, safety, patents, and other historical information.
- WHO International Clinical Trials Registry Platform (ICTRP) and ClinicalTrials.gov– the most comprehensive lists of global clinical trials available. Clinical trials will be scoped for relevance (defined as an investigation of one or more candidates or products with a primary and/or secondary outcome measure matching the scope). This step serves a dual function of uncovering additional candidates for inclusion, as well as capturing and linking clinical trial data to candidates marked for inclusion in the database (see Step 2 below for more information).
- EMBASE– for relevant product development literature to validate already identified and uncover new candidates for inclusion.
- Grant databases of relevant global funders of global health R&D – to validate existing and find new candidates (particularly those in preclinical/discovery stage). These include the United States National Institutes of Health (US NIH)’s RePORTER; the European Union/Commission’s CORDIS; the Gates Foundation (GF)’s grants database (supplied from the GF); SNSF, UKRI, and others as appropriate.
- Impact Global Health's G-FINDER R&D funding database for relevant projects. The SRH conditions covered in the pipeline have been included in the G-FINDER survey this year (2024) and the related grants will be included.
Step 2: Linking preclinical and clinical development data
Clinical trial data was collected via clinicaltrials.gov, WHO International Clinical Trials Registry Platform (ICTRP), and Adis Insight clinical trials outputs, linked to a diversity of clinical trial databases. We cross-checked interventions with the candidates identified in step 1, to provide more information on candidate clinical development. We cross-checked/follow up these outputs with relevant national clinical trials registers, as needed.
Step 3: Completing candidate and product profiles.
Much of the candidate information needed to complete candidate profiles was provided through Steps 1 and 2. In addition, we utilised academic literature search engines to source greater detail and context for the candidates identified in Steps 1 and 2. Primarily, we searched EMBASE using the candidate name/s, and reviewed relevant literature retrieved (including that already sourced in Step 1) to verify and cross reference information as needed. Additional information was searched for via relevant regulatory websites, such as the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) as appropriate, as well as a number of reliable online sources, including DRUGBANK Online, PubChem, the US National Library of Medicine’s Medical Subject Headings (MeSH) portal, and other websites as needed. Technical information on products was also sourced from developer websites and if applicable and available, product pamphlets/inserts.
Search terms & strategy for pipeline expansion
For most databases, we utilised variations and combinations of related search words – such as ‘gonorrhoeae’ OR ‘gonorr*’ – along with any timeframe filters to narrow datasets for review.
Table 3: ‘Condition’ key words for SRH search queries

For very large datasets – such as /EMBASE – our search strategy was to link three to four keywords together, and query these in all possible combinations. This included the ‘condition’ key words (as above, e.g. ‘gonorrhea’, etc.), combined with those related to ‘product type’ (‘drug’, ‘vaccine’, ‘diagnostic’, etc.), and to ‘research & development’ (‘innovation’, ‘discovery’, ‘preclinical’, ‘novel’, ‘clinical development’, etc.).
I.e.: ‘condition”’ + ‘product type’ + ’R&D’
(e.g., ‘gonorrhea’ + ‘drug’ + ‘novel’)
Table 4: 'Product type' and 'R&D' related key words for pipeline search queries
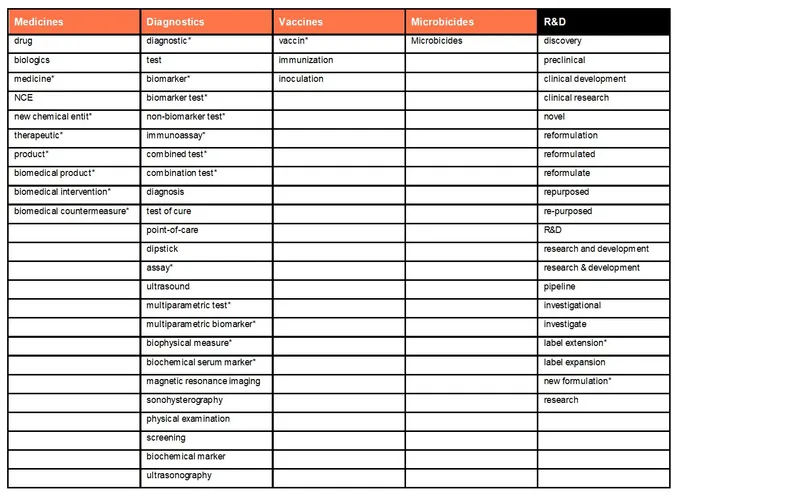
Table 1: Scope of pipeline

The pipeline we developed is candidate-focused (as opposed to clinical trial-focused) presenting a profile for each candidate identified. It focuses on active candidates, with evidence of activity between 2021 and 2024, and products (marketed, approved or adopted in clinical practice as gold standard – e.g. diagnostic tests).
This first iteration of the SRH pipeline focuses on sexually transmitted infections (STIs) with a risk of antimicrobial resistance (AMR). Based on the WHO’s evaluation, three STI pathogens are at risk of AMR: Neisseria gonorrhoeae, Mycoplasma genitalium and Trichomonas vaginalis.
The product categories included in the pipeline for these three STIs are drugs, biologics, vaccines, microbicides and diagnostics.
For inclusion in the pipeline dataset, medicine entries must:
- be drugs (small molecules intended for use in the cure, mitigation, treatment, or prevention of disease), biologics (animal or non-animal derived, immunoglobulins or non-immunoglobulin products) with no restrictions: they can be entirely new entities; existing/repurposed/label extensions; new formulations or dosing of existing/registered medicines.
- be preventive or therapeutic in nature – used to prevent the transmission of STIs, the acquisition/development of the infection or to eradicate the infection;
- have one or multiple indications related to either gonorrhoea, mycoplasma genitalium or trichomoniasis;
- either be in active discovery, pre-clinical or clinical development since 2021, or be approved and registered for clinical use and/or be used currently in clinical treatment (off-label), and;
- be applicable for use in LMIC context.
For inclusion in the pipeline dataset, diagnostic entries must:
- be predictive/screening and/or diagnostic tests, with no restrictions: biochemical serum markers; non-biomarker tests; or fixed multiparametric tests;
- have an indication or multiple indications related to gonorrhoea, mycoplasma genitalium or trichomoniasis; either for the diagnosis of the STI or for diagnosing antimicrobial resistance
- either be in active discovery, pre-clinical or clinical development since 2021, or be approved and registered for clinical use and/or be used currently in clinical treatment (off-label), and;
- be applicable for use in LMIC context.
For inclusion in the pipeline dataset, vaccines entries must:
- be preventive or therapeutic in nature – used to prevent the transmission of STIs, the acquisition/development of the infection or to clear the infection;
- have one or multiple indications related to either gonorrhoea, mycoplasma genitalium or trichomoniasis
- either be in active discovery, pre-clinical or clinical development since 2021, or be approved and registered for clinical use and/or be used currently in clinical treatment (off-label), and;
- be applicable for use in LMIC context.
For inclusion in the pipeline dataset, microbicide entries must:
- be topical microbicides intended to prevent transmission of N. gonorrhoea, M. genitalium or trichomonas vaginalis
- be suitable for use in LMICs (e.g. heat stable, easy to use)
- If microbicides are designed to prevent two or more STIs, or combined with contraceptives, they are considered multi-purpose preventive technologies (MPTs)
Exclusions:
- Dietary supplements are excluded;
- Predictive/diagnostic strategies, algorithms with no identifiable products; or that include risk models that comprise of multiple individual parameters/tests as a single candidate entry; (for example several individual biomarkers which diagnostic utility is evaluated separately and not as a combined test usable in the clinic);
- Clinical guidelines or criteria that are not a biomedical product are excluded.
The pipeline includes the following conditions and products:
- Gonorrhoea, Mycoplasma Genitalium and Trichomoniasis: drugs, biologics, vaccines, microbicides, diagnostics
All entries contain the same information. Data fields for each new candidate include all the following fields, where available and verifiable:
Table 2: Data fields

Data sources and approach
To build out comprehensive, candidate-driven pipeline datasets, we undertook a series of partially sequential, mutually reinforcing steps to build a picture of the candidates themselves, as well as the product development landscape for that SRH condition. Broadly (with more detail below), these steps are: (1) identify all candidates that are marketed or in development for each SRH health area and associated product type (e.g. Endometriosis diagnostics) and establish unique profiles for each with standardised classifications across a range of standard characteristics (2) gather information on that candidate’s clinical development and pre-clinical/clinical trial results, (3) research additional context around the product (e.g. development history, stakeholders, etc.), and (4) sense check our findings.
Step 1: Initial candidate and product identification.
We utilised various sources to uncover, identify and cross-check candidates or marketed products. Key sources include:
- Adis Insight– a leading medicines development database, which will give a comprehensive output of relevant pipeline drugs, biologics (and possibly devices) related to relevant SRH conditions. Adis Insight also returns detailed information on candidate deals, trials, safety, patents, and other historical information.
- WHO International Clinical Trials Registry Platform (ICTRP) and ClinicalTrials.gov– the most comprehensive lists of global clinical trials available. Clinical trials will be scoped for relevance (defined as an investigation of one or more candidates or products with a primary and/or secondary outcome measure matching the scope). This step serves a dual function of uncovering additional candidates for inclusion, as well as capturing and linking clinical trial data to candidates marked for inclusion in the database (see Step 2 below for more information).
- EMBASE– for relevant product development literature to validate already identified and uncover new candidates for inclusion.
- Grant databases of relevant global funders of global health R&D – to validate existing and find new candidates (particularly those in preclinical/discovery stage). These include the United States National Institutes of Health (US NIH)’s RePORTER; the European Union/Commission’s CORDIS; the Gates Foundation (GF)’s grants database (supplied from the GF); SNSF, UKRI, and others as appropriate.
- Impact Global Health's G-FINDER R&D funding database for relevant projects. The SRH conditions covered in the pipeline have been included in the G-FINDER survey this year (2024) and the related grants will be included.
Step 2: Linking preclinical and clinical development data
Clinical trial data was collected via clinicaltrials.gov, WHO International Clinical Trials Registry Platform (ICTRP), and Adis Insight clinical trials outputs, linked to a diversity of clinical trial databases. We cross-checked interventions with the candidates identified in step 1, to provide more information on candidate clinical development. We cross-checked/follow up these outputs with relevant national clinical trials registers, as needed.
Step 3: Completing candidate and product profiles.
Much of the candidate information needed to complete candidate profiles was provided through Steps 1 and 2. In addition, we utilised academic literature search engines to source greater detail and context for the candidates identified in Steps 1 and 2. Primarily, we searched EMBASE using the candidate name/s, and reviewed relevant literature retrieved (including that already sourced in Step 1) to verify and cross reference information as needed. Additional information was searched for via relevant regulatory websites, such as the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) as appropriate, as well as a number of reliable online sources, including DRUGBANK Online, PubChem, the US National Library of Medicine’s Medical Subject Headings (MeSH) portal, and other websites as needed. Technical information on products was also sourced from developer websites and if applicable and available, product pamphlets/inserts.
Search terms & strategy for pipeline expansion
For most databases, we utilised variations and combinations of related search words – such as ‘gonorrhoeae’ OR ‘gonorr*’ – along with any timeframe filters to narrow datasets for review.
Table 3: ‘Condition’ key words for SRH search queries

For very large datasets – such as /EMBASE – our search strategy was to link three to four keywords together, and query these in all possible combinations. This included the ‘condition’ key words (as above, e.g. ‘gonorrhea’, etc.), combined with those related to ‘product type’ (‘drug’, ‘vaccine’, ‘diagnostic’, etc.), and to ‘research & development’ (‘innovation’, ‘discovery’, ‘preclinical’, ‘novel’, ‘clinical development’, etc.).
I.e.: ‘condition”’ + ‘product type’ + ’R&D’
(e.g., ‘gonorrhea’ + ‘drug’ + ‘novel’)
Table 4: 'Product type' and 'R&D' related key words for pipeline search queries

Related insights & reports

45 min read
A New Era? Funding for sexual & reproductive health R&D 2018-2023
This is the fourth in a series of reports summarising the state of global funding for LMIC-applicable biomedical research and development targeting a range of sexual & reproductive health and women’s health issues


20 min read
European Investment in Sexual & Reproductive Health R&D: Steady Overall but Room to Grow
This snapshot offers insights into European R&D funding from 2018 to 2022 for sexual & reproductive health issues.


45 min read
G-FINDER 2023 Sexual & Reproductive Health Research & Development report: Beyond Spillovers
The 2023 report on investment in R&D for reproductive, sexual, maternal or women's health issues produced through the G-FINDER project
